scab: single cell analysis of B cells#
scab provides a computational framework for integrating, analyzing and visualizing single B cell multi-omics data. The scab package consists of two primary components:
- scabranger
a command line interface (CLI) wrapper around CellRanger that allows customizable batch processing of multiple samples with data spread across one or more sequencing runs
- scab API
a Python API for processing, analyzing, and visualizing single cell datasets, with a focus on B cell receptor (BCR) repertoire analysis
We have tried to design a simple, straightforward API that should feel quite familiar to users of scanpy. scab allows sophisticated analyses with just a few lines of code:
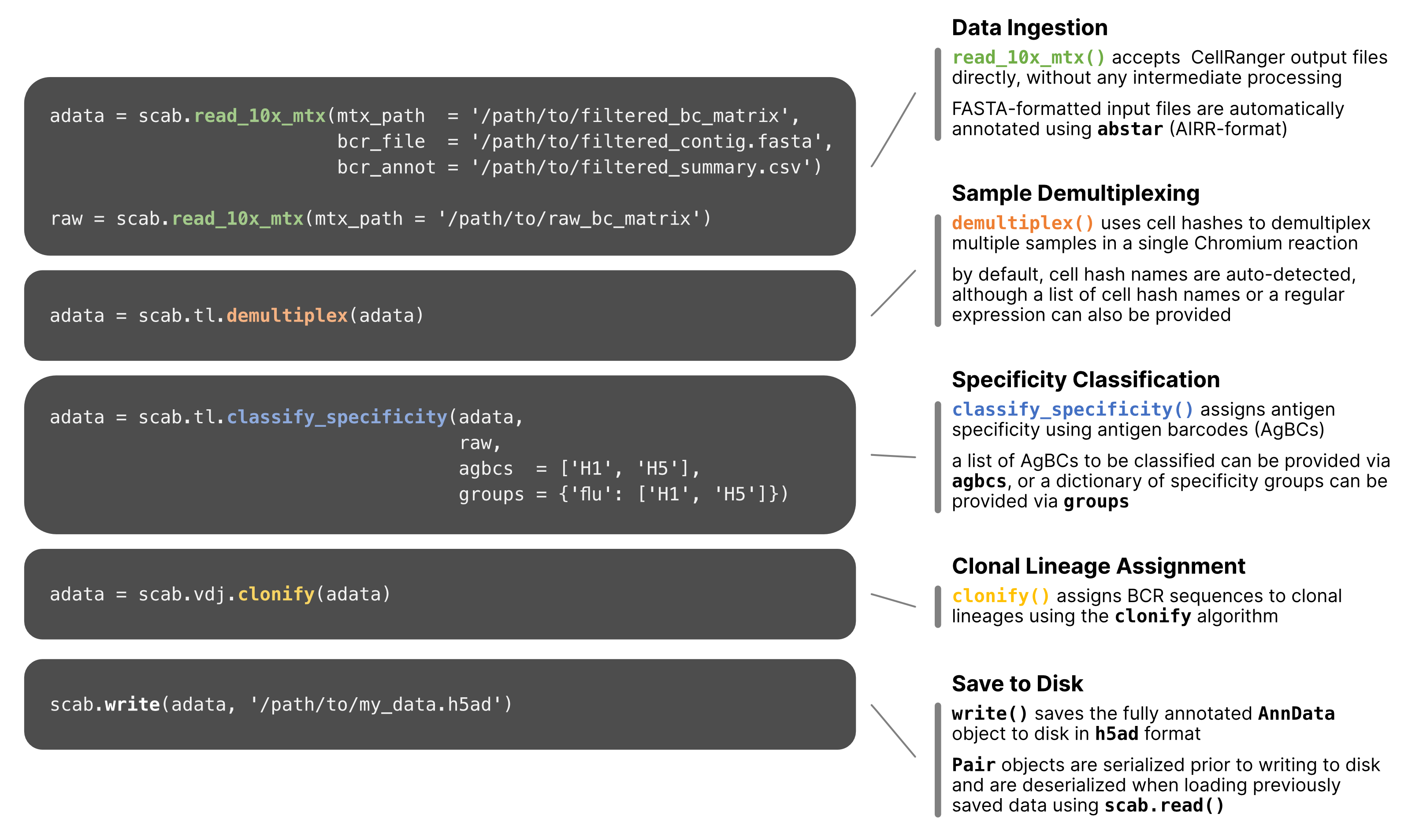

scab is a standards-based toolkit built on the AnnData objects used
by scanpy. We integrate several tools specifically designed to facilitate
analysis of adaptive immune cells and receptor repertoires.